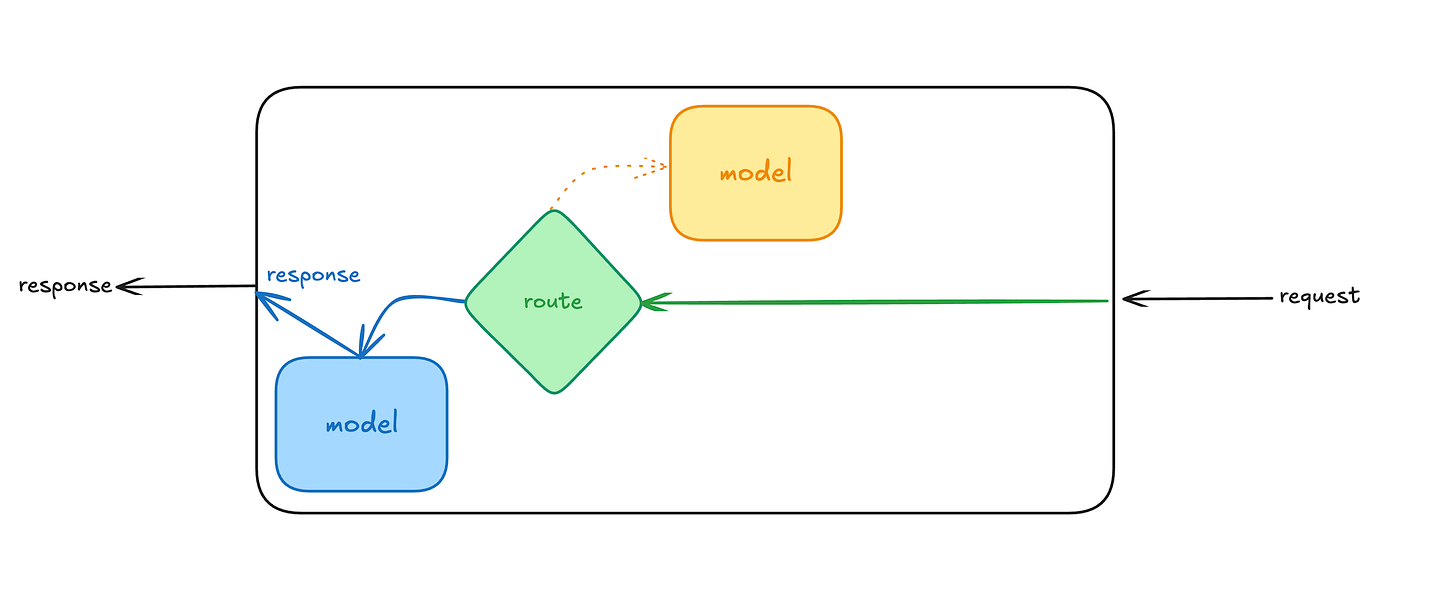
Juggling multiple models in a single serving endpoint
How to serve multiple models on a single model serving endpoint in Databricks using pyfunc
Have you ever found yourself juggling multiple ML models? Imagine this: you're maintaining a prediction service that started with a single model, but now you've got a dozen micro-models serving different business needs. Costs are climbing. You are dreaming of consolidation.
For most scenarios, Databricks Model Serving provides an easy solution. They allow you to deploy multiple models behind a single endpoint, split traffic, and route requests. This approach is perfect for A/B testing and canary deployments, where simple traffic splitting is sufficient. However, there are situations where we can hit limitations:
routing based on requests (e.g., user attributes)
routing based on time
managing dozens of micro-models and want to consolidate infrastructure
routing dynamically based on business rules
You could spin up separate endpoints for each, but that means more DBUs, more management overhead, etc. This is where creating a custom PyFunc wrapper can provide a solution. Note that this should be viewed as an edge case and not a default pattern.
Before diving into the implementation, let’s consider the limitations.
Individual model metrics are combined, so monitoring is more difficult.
Models are loaded together, so there could be a resource inefficiency.
Routing rules may obscure decision paths.
Model versioning is less transparent.
In this deep dive, we will explore a really simple pattern to help solve this issue using PyFunc. By creating a wrapper with PyFunc, we will package various models in one deployable artifact, implement routing logic to direct requests to the right model, and maintain an entry point.
Let’s quickly create some base models.
We are training two separate models using the same California Housing dataset. Because both models have the exact same input data schema and the expected output schema, we can expect that the Model Signature for both models will be the same.
import mlflow
import pandas as pd
import numpy as np
from sklearn.datasets import fetch_california_housing
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.ensemble import RandomForestRegressor
data = fetch_california_housing()
california_housing = pd.DataFrame(data.data, columns=data.feature_names)
california_housing['target'] = data.target
X_train, X_test, y_train, y_test = train_test_split(
california_housing.drop('target', axis=1),
california_housing['target'],
test_size=0.2,
random_state=42
)
lr_model = LinearRegression().fit(X_train, y_train)
rf_model = RandomForestRegressor().fit(X_train, y_train)
signature = mlflow.models.infer_signature(X_train, lr_model.predict(X_train))This represents a standard model development workflow. This pattern builds on existing models and training processes rather than replacing them. In other words, you can adopt this pattern without too much disruption to your current workflows.
These can now be logged and registered in Unity Catalog. Nothing new here!
with mlflow.start_run(run_name="California Housing Models") as housing_run:
mlflow.sklearn.log_model(lr_model, "linear_regression_model", signature=signature)
mlflow.sklearn.log_model(rf_model, "random_forest_model", signature=signature)
mlflow.set_registry_uri("databricks-uc")
mlflow.register_model(
f"runs:/{housing_run.info.run_id}/linear_regression_model",
"your_catalog.your_schema.california_housing_linear_regression"
)
mlflow.register_model(
f"runs:/{housing_run.info.run_id}/random_forest_model",
"your_catalog.your_schema.california_housing_random_forest"
)
Create a custom model using pyfunc.
We are going to use pyfunc to orchestrate and serve as the main interface for interacting with the base models. The wrapper will load our models and dynamically select which model to use based on the request parameters.
class ModelRouter(mlflow.pyfunc.PythonModel):
def load_context(self, context):
self.linear_model = mlflow.sklearn.load_model(
context.artifacts["linear_regression_model"]
)
self.forest_model = mlflow.sklearn.load_model(
context.artifacts["random_forest_model"]
)
def predict(self, context, model_input):
# The 'model' column specifies which model to use
if model_input['model'].eq('RandomForest').any():
return {
"prediction": self.forest_model.predict(model_input.drop('model', axis=1))
}
elif model_input['model'].eq('LinearRegression').any():
return {
"prediction": self.linear_model.predict(model_input.drop('model', axis=1))
}
else:
raise ValueError("Unrecognized model type. Use 'RandomForest' or 'LinearRegression'")I want to highlight two important aspects of this wrapper. First, in load_context, we are loading the underlying Linear Regression and Random Forest models from the artifacts. When we log and register this wrapper, we will need to specify these artifacts, so that the wrapper will correctly load the models that we trained. Keep in mind that in the model serving environment, load_context is called once, so loading the models should not affect the serving latency after initialization.
Second, there is a lot of flexibility here. In the code snippet, we are using an extra column in the model input called model to select which model to use. But you can implement virtually any routing logic. You can switch between the models based on geographic location or the time the request was submitted.
Registering the wrapper with the model artifacts.
In order to register the model, we need to create a proper Model Signature. I am going to use the infer_signature function to do so. You can also manually construct the signature object. The signature will be similar to the signatures used for the base models. Because our wrapper uses an extra column to decide which model to use, we need to take that into consideration.
input_example = X_train.copy()
input_example['model'] = 'RandomForest'
router_signature = mlflow.models.infer_signature(
input_example,
{"prediction": rf_model.predict(X_train)}
)When we log the model, we need to include the base models as artifacts:
with mlflow.start_run() as run:
router_model = ModelRouter()
mlflow.pyfunc.log_model(
"model_router",
python_model=router_model,
signature=router_signature,
artifacts={
"linear_regression_model":
"models:/your_catalog.your_schema.california_housing_linear_regression/1",
"random_forest_model":
"models:/your_catalog.your_schema.california_housing_random_forest/1",
},
extra_pip_requirements=["scikit-learn==1.4.2", "numpy==1.23.5", "pandas==1.5.3"]
)
# Register the router model
mlflow.register_model(
f"runs:/{run.info.run_id}/model_router",
"your_catalog.your_schema.housing_model_router"
)Now, we have created a self-contained wrapper that includes everything needed for serving.
What happens when we are dealing with different inputs?
Imagine your system spans multiple domains. Different data, different tasks, but you still need a unified interface.
First, let’s train model on a different dataset.
from sklearn.datasets import load_breast_cancer
from sklearn.ensemble import RandomForestClassifier
cancer_data = load_breast_cancer()
cancer_df = pd.DataFrame(cancer_data.data, columns=cancer_data.feature_names)
cancer_df['target'] = cancer_data.target
X_train_cancer, X_test_cancer, y_train_cancer, y_test_cancer = train_test_split(
cancer_df.drop('target', axis=1),
cancer_df['target'],
test_size=0.2
)
cancer_model = RandomForestClassifier().fit(X_train_cancer, y_train_cancer)
with mlflow.start_run() as run:
cancer_signature = mlflow.models.infer_signature(
X_train_cancer,
cancer_model.predict(X_train_cancer)
)
mlflow.sklearn.log_model(
cancer_model,
"random_forest_cancer",
signature=cancer_signature
)
mlflow.register_model(
f"runs:/{run.info.run_id}/random_forest_cancer",
f"{catalog}.{db}.{rf_br_model_name}"
)Now, we can create a wrapper that handles both datasets. This wrapper is similar to the previous one.
class MultiDomainRouter(mlflow.pyfunc.PythonModel):
def load_context(self, context):
self.housing_model = mlflow.sklearn.load_model(
context.artifacts["housing_model"]
)
self.cancer_model = mlflow.sklearn.load_model(
context.artifacts["cancer_model"]
)
self.housing_columns = context.artifacts['housing_features']
self.cancer_columns = context.artifacts['breast_cancer_features']
def predict(self, context, model_input):
if model_input['domain'].eq('housing').any():
# validate input data
input_cols = set(model_input.columns) - {'domain'}
missing_cols = self.housing_columns - input_cols
if missing_cols:
raise ValueError(f"Missing required columns for model: {missing_cols}")
# columns needed by model
features = model_input[list(self.housing_columns) + ['domain']]
return {
"prediction": self.housing_model.predict(
features.drop('domain', axis=1)
)
}
elif model_input['domain'].eq('cancer').any():
# validate input data
input_cols = set(model_input.columns) - {'domain'}
missing_cols = self.cancer_columns - input_cols
if missing_cols:
raise ValueError(f"Missing required columns for model: {missing_cols}")
# columns needed by model
features = model_input[list(self.cancer_columns) + ['domain']]
return {
"prediction": self.cancer_model.predict(
features.drop('domain', axis=1)
)
}
else:
raise ValueError("Unrecognized domain. Use 'housing' or 'cancer'")But wait— how are we supposed to define the Model Signature? The expected inputs will be different.
A quick aside on Model Signatures.
A Model Signature defines the input and output schema that the model is expected to receive and output. There are two main types of signatures: column-based (used for most traditional ML models) and tensor-based (used for deep learning applications).
Column-based signatures consist of a list of columns (very surprising), each with an expected data type. The signature for the California Housing models look like this:
inputs:
['MedInc': double (required), 'HouseAge': double (required), 'AveRooms': double (required), 'AveBedrms': double (required), 'Population': double (required), 'AveOccup': double (required), 'Latitude': double (required), 'Longitude': double (required)]
outputs:
[Tensor('float64', (-1,))]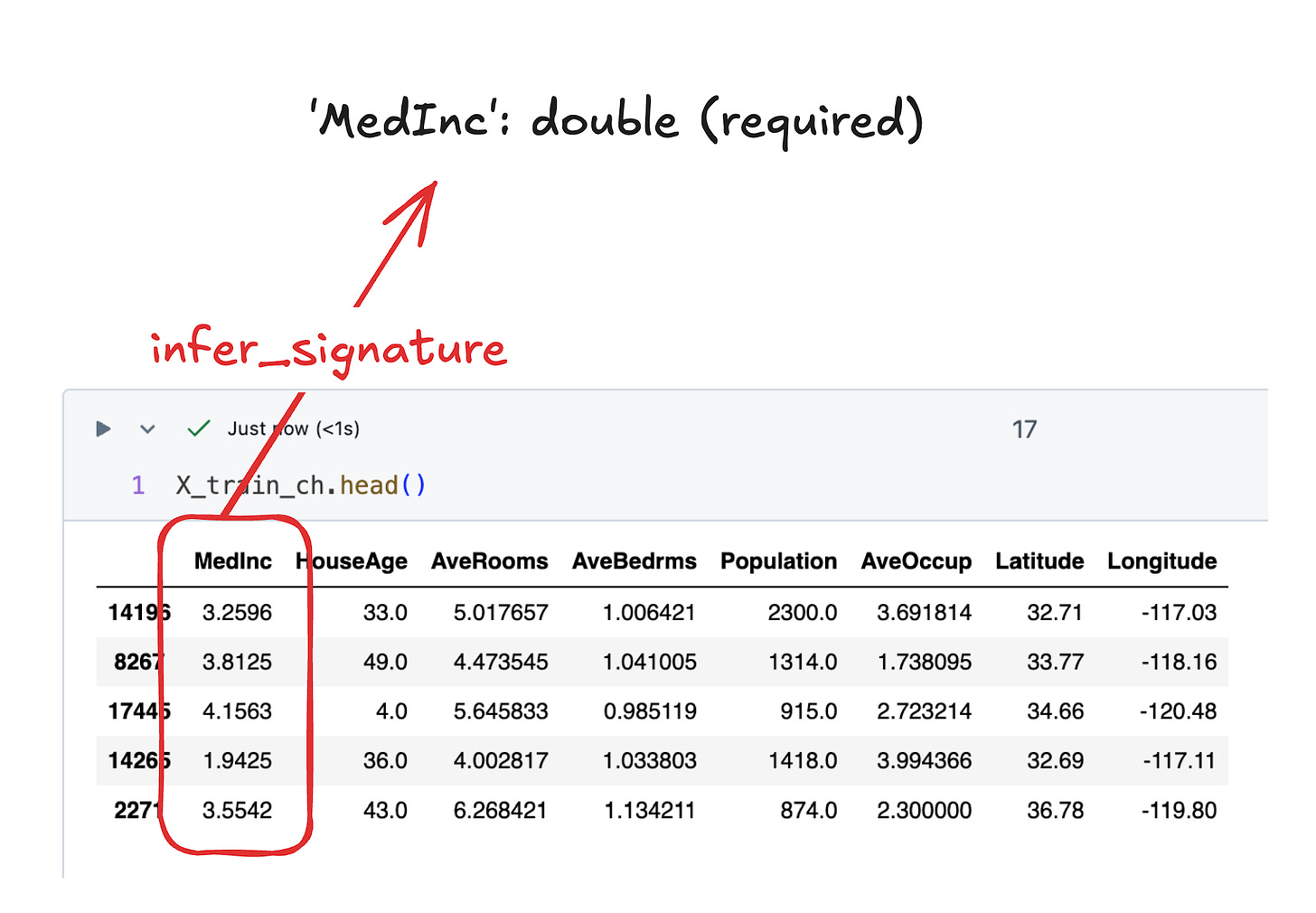
Notice: each of these columns are required by the model. Required fields must be included in the input, and if it is not there, it will error out. However, we can include optional fields as well.
The function assumes that these columns are required because in the dataframe I used, all values were properly populated. In order to configure a field as optional, we can use mlflow.models.infer_signature by passing in some None values for that field. Basically, we can concat the two datasets and infer the signature to make all of the columns for both types of inputs optional.
bc_head = bc.head()
california_housing_head = california_housing.head()
merged_df = pd.concat([bc_head, california_housing_head])
merged_df_output = {"california_housing": rf_ch.predict(california_housing_head.drop('target', axis=1))}
signature_merged_df = infer_signature(merged_df, merged_df_output)By making most fields optional in the signature, we're essentially telling MLflow to let all requests through to our code. We will still need to do the validation for each model. This gives us the flexibility to route between completely different models while still maintaining a consistent interface for client applications.
If you look at the code snippet where we defined the model wrapper again, you can see that we have performed the validation of the model inputs ourselves. We have manually added feature names that are required for each model to enforce the input schema.
Conclusion
Voilà! This approach should address more complex needs to routing logic and consolidation of small models. It is perfect when you need:
request-level routing beyond percentage-based traffic splitting
multiple small models where separate endpoints would be inefficient.
complex routing logic based on request properties.
This router pattern gives you flexibility way to consolidate multiple models behind a single endpoint. Remember to think about the limitations we listed out earlier before implementing this in production!
Happy wrapping!